qtseries
qtseries.Rmd
library(timeseries)
library(gapminder)
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, unionThis is how the function qtseries works:
Building a time series plot with a dummy data set:
testdf <- data.frame(year = rep(seq(from = 1980, to= 1990, 1), 3),
rainfall = c(rnorm(11, 100, 30), rnorm(11, 70, 10),rnorm(11, 20, 20)),
temp = c(rnorm(11, 70, 30), rnorm(11, 100, 10),rnorm(11, 45, 20)),
daysofsun = c(NA, NA, rnorm(9, 280, 50), rnorm(11, 300, 30), NA, rnorm(10, 340, 15)),
region = c(rep("A",11), rep("B", 11), rep("C", 11)))
qtseries(testdf, year, daysofsun, region, verbose = TRUE)
#> Plotting time series...Done labelling plot!...Done applying default theme aesthetics!
#> Warning: Removed 3 rows containing missing values (geom_point).
#> Warning: Removed 3 row(s) containing missing values (geom_path).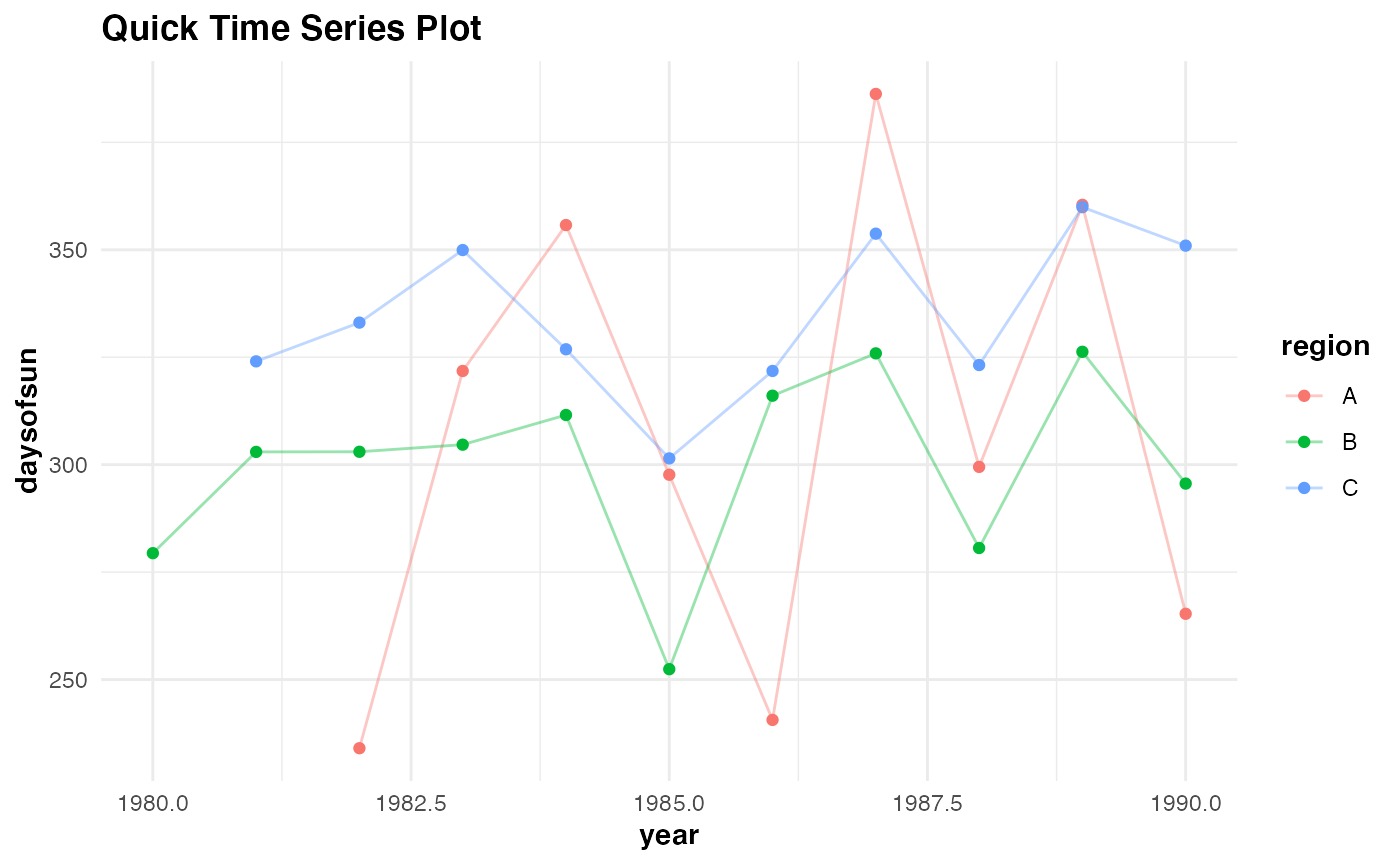
Building a time series plot with the gapminder data set:
qtseries(gapminder %>%
filter(continent == "Europe"), year, pop, country, verbose = TRUE)
#> Plotting time series...Done labelling plot!...Done applying default theme aesthetics!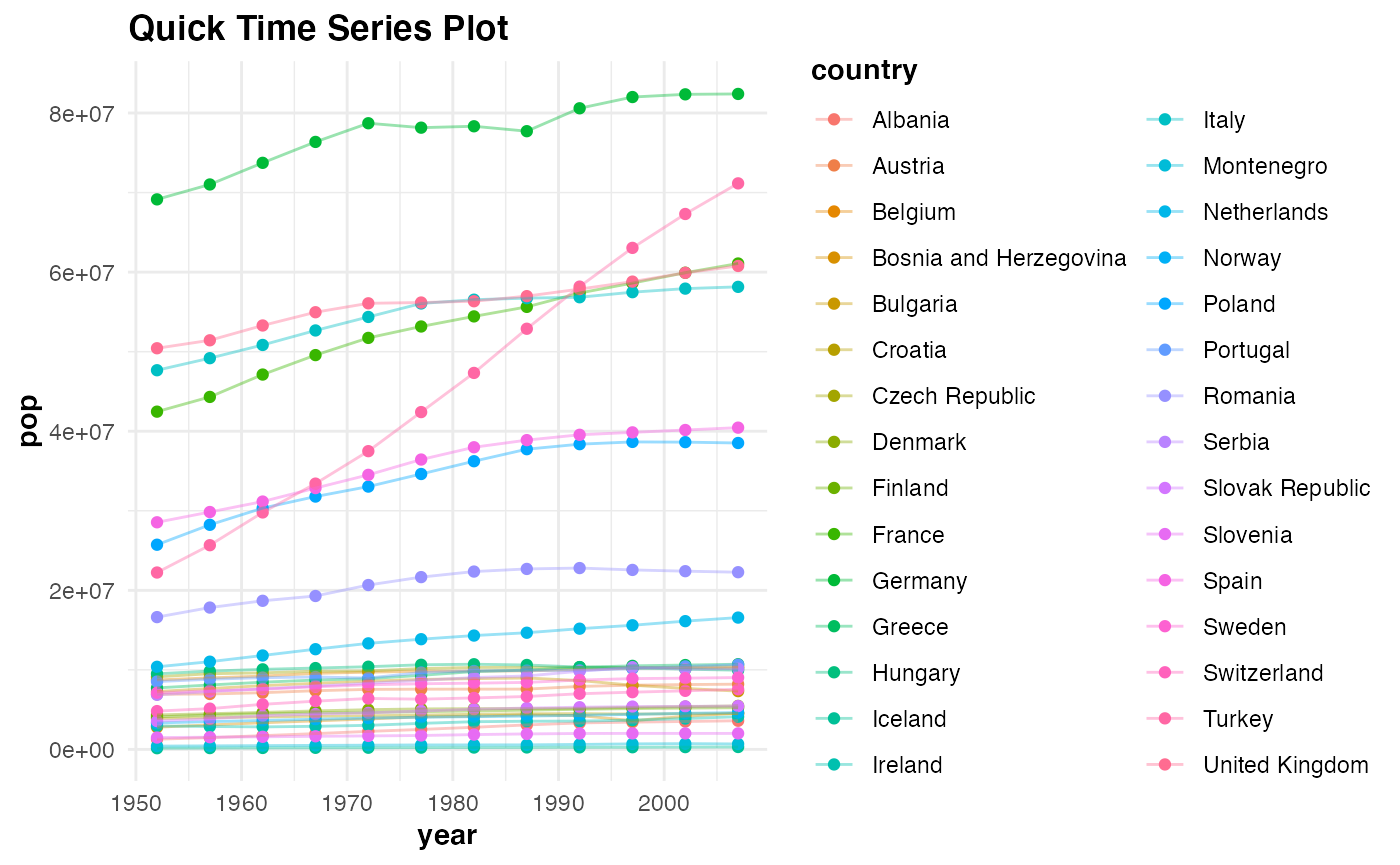
Some errors you might see if qtseries is given non-numeric variables for the time or voi inputs:
qtseries(gapminder %>%
filter(continent == "Americas"),continent, pop, country, verbose = TRUE)
#> Error in qtseries(gapminder %>% filter(continent == "Americas"), continent, : Sorry, the time variable needs to be a numeric variable!
qtseries(gapminder %>%
filter(continent == "Europe"),year, country, continent, verbose = TRUE)
#> Error in qtseries(gapminder %>% filter(continent == "Europe"), year, country, : Sorry, the variable of interest (dependent) needs to be a numeric variable!